PEPTIDES AND PROTEINS
Peptide Bond:
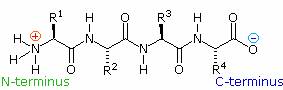
If the amine and carboxylic acid functional groups in amino acids join together to form amide bonds, a chain of amino acid units, called a peptide, is formed. A simple tetrapeptide structure is shown in the following diagram. By convention, the amino acid component retaining a free amine group is drawn at the left end (the N-terminus) of the peptide chain, and the amino acid retaining a free carboxylic acid is drawn on the right (the C-terminus). As expected, the free amine and carboxylic acid functions on a peptide chain form a zwitterionic structure at their isoelectric pH.


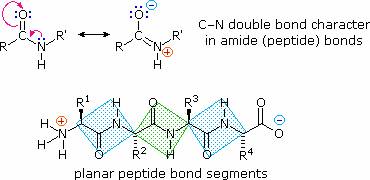
The conformational flexibility of peptide chains is limited chiefly to rotations about the bonds leading to the alpha-carbon atoms. This restriction is due to the rigid nature of the amide (peptide) bond. As shown in the following diagram, nitrogen electron pair delocalization into the carbonyl group results in significant double bond character between the carbonyl carbon and the nitrogen. This keeps the peptide links relatively planar and resistant to conformational change. The color shaded rectangles in the lower structure define these regions, and identify the relatively facile rotations that may take place where the corners meet (i.e. at the alpha-carbon). This aspect of peptide structure is an important factor influencing the conformations adopted by proteins and large peptides.
PEPTIDES:
Because the N-terminus of a peptide chain is distict from the C-terminus, a small peptide composed of different aminoacids may have a several constitutional isomers. For example, a dipeptide made from two different amino acids may have two different structures. Thus, aspartic acid (Asp) and phenylalanine (Phe) may be combined to make Asp-Phe or Phe-Asp, remember that the amino acid on the left is the N-terminus. The methyl ester of the first dipeptide (structure on the right) is the artificial sweetner aspartame, which is nearly 200 times sweeter than sucrose. Neither of the component amino acids is sweet (Phe is actually bitter), and derivatives of the other dipeptide (Phe-Asp) are not sweet.

A tripeptide composed of three different amino acids can be made in 6 different constitutions, and the tetrapeptide shown above (composed of four different amino acids) would have 24 constitutional isomers. When all twenty of the natural amino acids are possible components of a peptide, the possible combinations are enormous. Simple statistical probability indicates that the decapeptides made up from all possible combinations of these amino acids would total 2010!.
Natural peptides of varying complexity are abundant. The simple and widely distributed tripeptide glutathione (first entry in the following table), is interesting because the side-chain carboxyl function of the N-terminal glutamic acid is used for the peptide bond. An N-terminal glutamic acid may also close to a lactam ring, as in the case of TRH (second entry). The abbreviation for this transformed unit is pGlu (or pE), where p stands for "pyro" (such ring closures often occur on heating). The larger peptides in the table also demonstrate the importance of amino acid abbreviations, since a full structural formula for a nonapeptide (or larger) would prove to be complex and unwieldy. The formulas using single letter abbreviations are colored red. The ten peptides listed in this table make use of all twenty common amino acids. Note that the C-terminal unit has the form of an amide in some cases (e.g. TRH, angiotensin & oxytocin). When two or more cysteines are present in a peptide chain, they are often joined by disulfide bonds (e.g. oxytocin & endothelin); and in the case of insulin, two separate peptide chains (A & B) are held together by such links.

PRIMARY STRUCTURE ANALYSIS:
The different amino acids that
make up a peptide or protein, and the order in which they are joined together by
peptide bonds is referred to as the primary structure. From the examples shown
above, it should be evident that it is not a trivial task to determine the
primary structure of such compounds, even modestly sized ones.
Complete hydrolysis of a protein or peptide, followed by amino acid analysis
establishes its gross composition, but does not provide any bonding sequence
information.

Partial hydrolysis will produce a mixture of shorter peptides and some amino acids. If the primary structures of these fragments are known, it is sometimes possible to deduce part or all of the original structure by taking advantage of overlapping pieces. For example, if a heptapeptide was composed of three glycines, two alanines, a leucine and a valine, many possible primary structures could be written. On the other hand, if partial hydrolysis gave two known tripeptide and two known dipeptide fragments, as shown on the right, simple analysis of the overlapping units identifies the original primary structure. Of course, this kind of structure determination is very inefficient and unreliable. First, we need to know the structures of all the overlapping fragments. Second, larger peptides would give complex mixtures which would have to be separated and painstakingly examined to find suitable pieces for overlapping. It should be noted, however, that modern mass spectrometry uses this overlap technique effectively. The difference is that bond cleavage is not achieved by hydrolysis, and computers assume the time consuming task of comparing a multitude of fragments.
N-Terminal group Analysis:

Over the years that chemists have been studying these important natural products, many techniques have been used to investigate their primary structure or amino acid sequence. Indeed, commercial instruments that automatically sequence peptides and proteins are now available. A few of the most important and commonly used techniques will be described here. Identification of the N-terminal and C-terminal aminoacid units of a peptide chain provides helpful information. N-terminal analysis is accomplished by the Edman Degradation, which is outlined in the following diagram. A free amine function, usually in equilibrium with zwitterion species, is necessary for the initial bonding to the phenyl isothiocyanate reagent. The products of the Edman degradation are a thiohydantoin heterocycle incorporating the N-terminal amino acid together with a shortened peptide chain. Amine functions on a side-chain, as in lysine, may react with the isothiocyanate reagent, but do not give thiohydantoin products. A major advantage of the Edman procedure is that the remaining peptide chain is not further degraded by the reaction. This means that the N-terminal analysis may be repeated several times, thus providing the sequence of the first three to five amino acids in the chain. A disadvantage of the procedure is that is peptides larger than 30 to 40 units do not give reliable results.
C-Terminal group Analysis:
Chemical Analysis:
Complementary C-terminal analysis of peptide chains may be accomplished chemically or enzymatically. The chemical analysis is slightly more complex than the Edman procedure. First, side-chain carboxyl groups and hydroxyl groups must be protected as amides or esters. Next, the C-terminal carboxyl group is activated as an anhydride and reacted with thiocyanate. The resulting acyl thiocyanate immediately cyclizes to a hydantoin ring, and this can be cleaved from the peptide chain in several ways, not described here. Depending on the nature of this final cleavage, the procedure can be modified to give a C-terminal acyl thiocyanate peptide product which automatically rearranges to a thiohydantoin incorporating the penultimate C-terminal unit. Thus, repetitive analyses may be conducted in much the same way they are with the Edman procedure.

Enzyme Analysis:
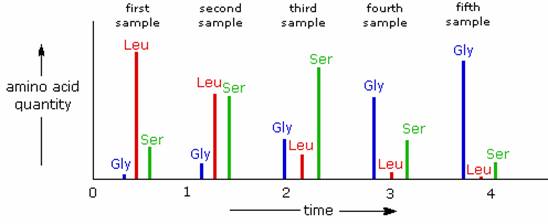
Enzymatic C-terminal amino acid cleavage by one of several carboxypeptidase enzymes is a fast and convenient method of analysis. Because the shortened peptide product is also subject to enzymatic cleavage, care must be taken to control the conditions of reaction so that the products of successive cleavages are properly monitored. The following example illustrates this feature. A peptide having a C-terminal sequence: ~Gly-Ser-Leu is subjected to carboxypeptidase cleavage, and the free aminoacids cleaved in this reaction are analyzed at increasing time intervals. The leucine is cleaved first, the serine second, and the glycine third, as demonstrated by the sequential analysis. Of course, fourth and fifth units will also be released as time passes.
Peptide Hydrolysis (Cleavage):
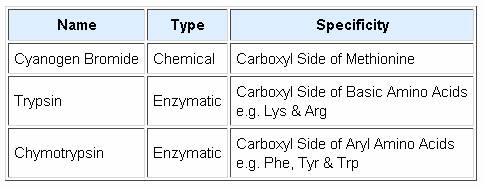
Since end group analysis of large peptides and proteins is of limited value, methods of selectively cleaving such macromolecules into smaller peptide fragments are commonly employed as a major step in structure elucidation. Three selective cleavage methods are outlined in the table on the left. These procedures all cleave peptide chains at designated locations, and at the carboxyl side of the targeted amino acid. A plausible mechanism for the cyanogen bromide cleavage is outlined below. The C-terminal side of the methionine is obtained as a smaller peptide, which can be examined by any of the preceding techniques. The N-terminal side is characterized by a homoserine lactone at its C-terminus. Mechanisms for the enzymatic reactions are not as easily formulated. Other enzymatic cleavages have been developed, but the two listed here will serve to illustrate their application.
Cyclic Peptides:
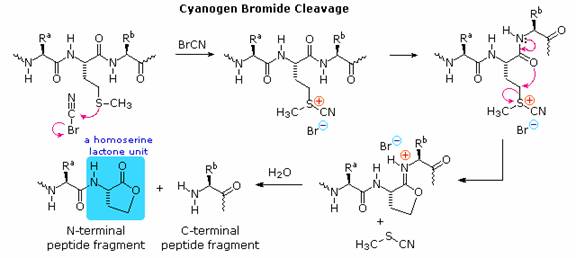
If the carboxyl function at the C-terminus of a peptide forms a peptide bond with the N-terminal amine group a cyclic peptide is formed. Carboxyate and amine functions on side chains may also combine to form rings. Cyclic peptides are most commonly found in microorganisms, and often incorporate some D-amino acids as well as unusual amino acids such as ornithine (Orn). The decapeptide antibiotic gramacidin S, produced by a strain of Bacillus brevis, is one example of this interesting class of natural products. The structure of gramicidin S is shown in the following diagram. The atypical amino acids are colored. When using a shorthand notation for cyclic structures, the top line is written by the usual convention (N-group on the left), but vertical and lower lines must be adjusted to fit the bonding. Arrows on these bonds point in the CO-N direction of each peptide bond.
SIMPLE PROTEINS:
Two general categories of simple proteins are commonly recognized.
|
S. No |
Type |
Description |
| 1. | Fibrous Proteins |
As the name implies, these substances have fibre-like structures, and serve
as the chief structural material in various tissues. Corresponding to this structural function, they are relatively insoluble in water and unaffected by moderate changes in temperature and pH. Subgroups within this category include: Collagens & Elastins, the proteins of connective tissues. tendons and ligaments. Keratins, proteins that are major components of skin, hair, feathers and horn. Fibrin, a protein formed when blood clots. |
| 2. | Globular Proteins |
Members of this class serve regulatory, maintenance and catalytic roles in
living organisms. They include hormones, antibodies and enzymes. and either dissolve or form colloidal suspensions in water. Such proteins are generally more sensitive to temperature and pH change than their fibrous counterparts. |
PEPTIDE SYNTHESIS:
In order to synthesize a peptide from its component amino acids, two obstacles must be overcome. The first of these is statistical in nature, and is illustrated by considering the dipeptide Ala-Gly as a proposed target. If we ignore the chemistry involved, a mixture of equal molar amounts of alanine and glycine would generate four different dipeptides. These are: Ala-Ala, Gly-Gly, Ala-Gly & Gly-Ala. In the case of tripeptides, the number of possible products from these two amino acids rises to eight. Clearly, some kind of selectivity must be exercised if complex mixtures are to be avoided. The second difficulty arises from the fact that carboxylic acids and 1º or 2º-amines do not form amide bonds on mixing, but will generally react by proton transfer to give salts.
Chemical Synthesis:
From the perspective of an organic chemist, peptide synthesis requires selective acylation of a free amine. To accomplish the desired amide bond formation, we must first deactivate all extraneous amine functions so they do not compete for the acylation reagent. Then we must selectively activate the designated carboxyl function so that it will acylate the one remaining free amine. Fortunately, chemical reactions that permit us to accomplish these selections are well known.
First, the basicity and
nucleophilicity of amines are substantially reduced by amide formation.
Consequently, the acylation of amino acids by treatment with acyl chlorides or
anhydrides at pH > 10, serves to protect their amino groups from further
reaction.
Second, acyl halide or anhydride-like activation of a specific carboxyl reactant must occur as a prelude to peptide (amide) bond formation. This is possible, provided competing reactions involving other carboxyl functions that might be present are precluded by preliminary ester formation. Remember, esters are weaker acylating reagents than either anhydrides or acyl halides.
Finally, dicyclohexylcarbodiimide (DCC) effects the dehydration of a carboxylic acid and amine mixture to the corresponding amide under relatively mild conditions. The strategy for peptide synthesis, as outlined here, should now be apparent. The following example shows a selective synthesis of the dipeptide Ala-Gly.

An Important issue remains to be addressed. Since the N-protective group is an amide, removal of this function might require conditions that would also cleave the just formed peptide bond. Furthermore, the harsh conditions often required for amide hydrolysis might cause extensive racemization of the amino acids in the resulting peptide. This problem strikes at the heart of our strategy, so it is important to give careful thought to the design of specific N-protective groups. In particular, three qualities are desired:
1) The protective amide should be easy to attach to amino acids.
2) The protected amino group should not react under peptide forming conditions.
3) The protective amide group should be easy to remove under mild conditions.
A number of protective groups that satisfy these conditions have been devised; and two of the most widely used, carbobenzoxy (Cbz) and t-butoxycarbonyl (BOC or t-BOC), are described here.
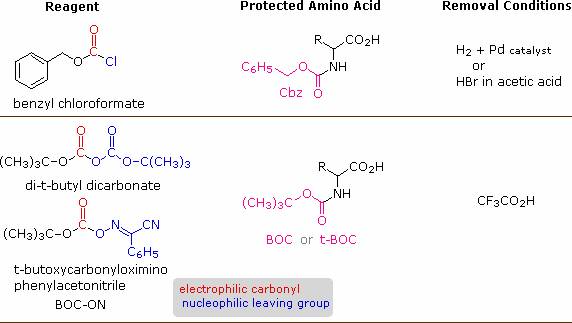
The reagents for introducing these N-protective groups are the acyl chlorides or anhydrides shown in the left portion of the above diagram. Reaction with a free amine function of an amino acid occurs rapidly to give the "protected" amino acid derivative shown in the center. This can then be used to form a peptide (amide) bond to a second amino acid. Once the desired peptide bond is created the protective group can be removed under relatively mild non-hydrolytic conditions.

Cleavage of the reactive benzyl or tert-butyl groups generates a common carbamic acid intermediate (HOCO-NHR) which spontaneously loses carbon dioxide, giving the corresponding amine. If the methyl ester at the C-terminus is left in place, this sequence of reactions may be repeated, using a different N-protected amino acid as the acylating reagent. Removal of the protective groups would then yield a specific tripeptide, determined by the nature of the reactants and order of the reactions.
Solid Phase Peptide Synthesis:
The synthesis of a peptide of significant length (e.g. ten residues) by this approach requires many steps, and the product must be carefully purified after each step to prevent unwanted cross-reactions. To facilitate the tedious and time consuming purifications, and reduce the material losses that occur in handling, a clever modification of this strategy has been developed. This procedure, known as the Merrifield Synthesis after its inventor R. Bruce Merrifield, involves attaching the C-terminus of the peptide chain to a polymeric solid, usually having the form of very small beads. Separation and purification is simply accomplished by filtering and washing the beads with appropriate solvents. The reagents for the next peptide bond addition are then added, and the purification steps repeated. The entire process can be automated, and peptide synthesis machines based on the Merrifield approach are commercially available. The final step, in which the completed peptide is released from the polymer support, is a simple benzyl ester cleavage.
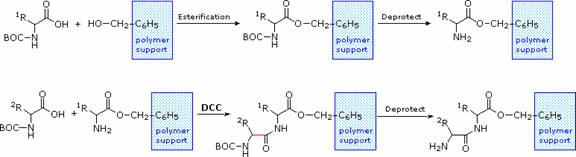
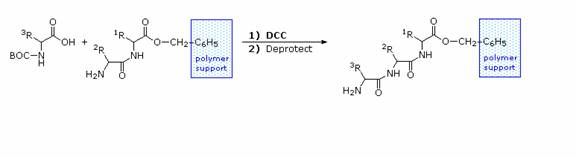
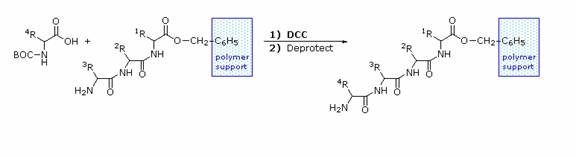
Two or more moderately sized peptides can be joined together by selective peptide bond formation, provided side-chain functions are protected and do not interfere. In this manner good sized peptides and small proteins may be synthesized in the laboratory. However, even if chemists assemble the primary structure of a natural protein in this or any other fashion, it may not immediately adopt its native secondary, tertiary and quaternary structure. Many factors, such as pH, temperature and inorganic ion concentration influence the conformational coiling of peptide chains. Indeed, scientists are still trying to understand how and why these higher structures are established in living organisms.
Denaturation:
The natural or native structures of proteins may be altered, and their biological activity changed or destroyed by treatment that does not disrupt the primary structure. This denaturation is often done deliberately in the course of separating and purifying proteins. For example, many soluble globular proteins precipitate if the pH of the solution is set at the pI of the protein. Also, addition of trichloroacetic acid or the bis-amide urea (NH2CONH2) is commonly used to effect protein precipitation. Following denaturation, some proteins will return to their native structures under proper conditions; but extreme conditions, such as strong heating, usually cause irreversible change.
