UM-BBD Pathway Prediction System (PPS) Tutorial
Mark Fischbach, John Schrom, Lynda Ellis,Jennifer Chiantello, Junfeng Gao, Michael TurnbullVersion 2.2 – February 14, 2008
Web version of this Tutorial:
http://umbbd.msi.umn.edu/predict/aboutPPS.html
OVERVIEW
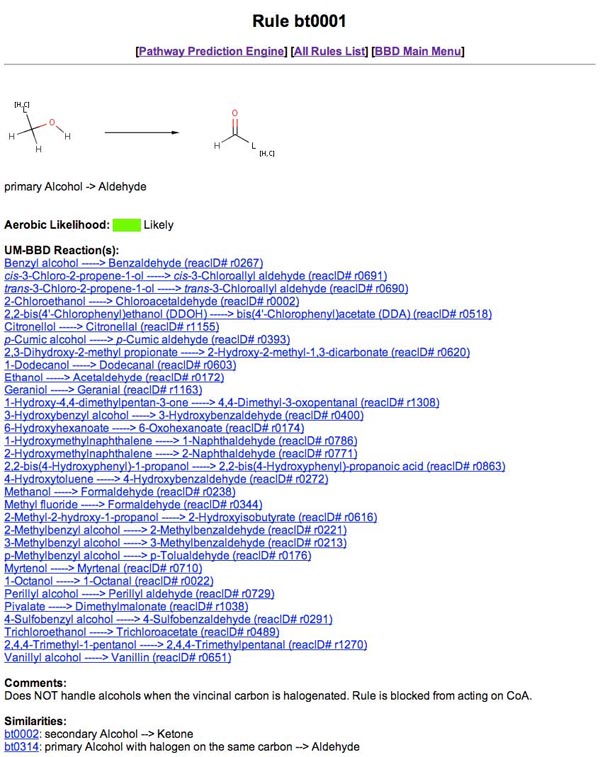
The UM-BBD Pathway Prediction System (PPS) predicts microbial catabolic reactions using substructure searching, a rule-base, and atom-to-atom mapping. The PPS recognizes organic functional groups found in a compound and predicts transformations based on biotransformation rules. These rules are based on reactions found in the UM-BBD database. The pathway prediction system can be accessed at the UM-BBD Pathway Prediction page, which can be reached from the "Pathway Prediction" link on the UM-BBD home page, or by using the URL: http://umbbd.msi.umn.edu/predict/. The PPS predicts biodegradation of user compounds. Users can choose if they will view all or only the more likely aerobic transformations. The PPS uses Chemaxon's MarvinSketch and MarvinView Java applets as plugins. A user enters a compound into the system by one of two methods: “Drawing the Structure and Generating SMILES” or “Entering SMILES Directly”.
DEMONSTRATION
Start the Demo
The PPS provides a demonstration of how the system works using the compound Benzyl Alcohol. To access the demo, click on the "Demo" button located under the MarvinSketch window of the UM-BBD Pathway Prediction Page.
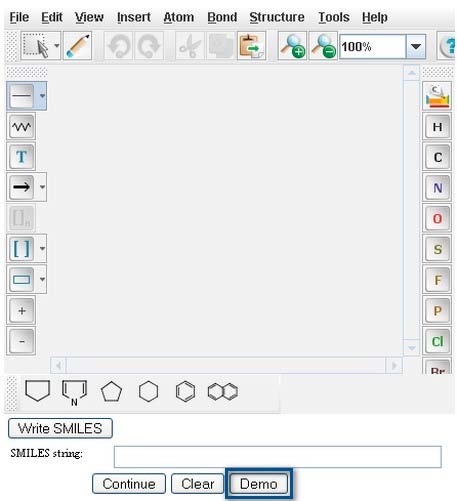
The Smiles string of Benzyl Alcohol will appear in the "Smiles string" box.
Pathway Prediction Results
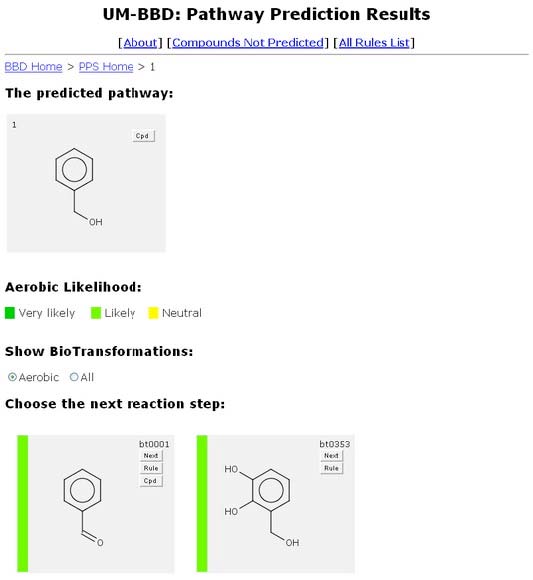
The system will automatically bring the user to the Pathway Prediction Results page for Benzyl Alcohol. The default se tting for the PPS is “Aerobic” which will show the biotransformations most likely to occur under normal conditions (exposed to air, in the top layers of or in well oxygenated soil or water). You can change this setting to “All” at any time, as described under Aerobic Likelihood below.
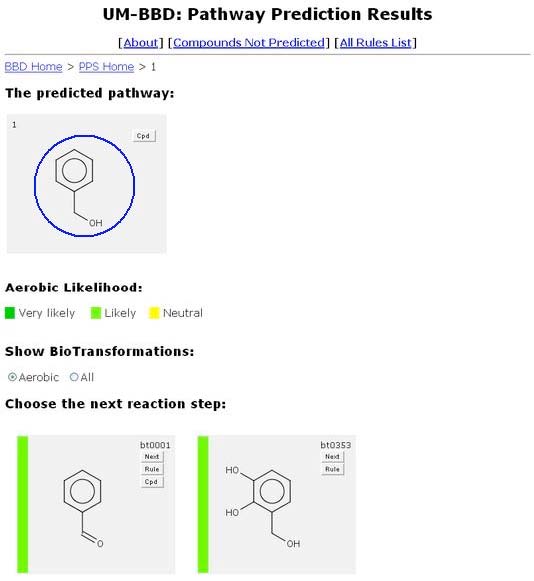
From the Pathway Prediction Results page, a user is able to see all the biotrans-formations of Benzyl Alcohol based on the rules found in the UM-BBD. The Navigation Bar “BBD Home > PPS Home > 1” permits quick navigation between different PPS prediction levels and the PPS and BBD home pages.
MarvinView
The user can click on any grey compound square, in either "The predicted pathway:", or"Choose the next reaction step:".
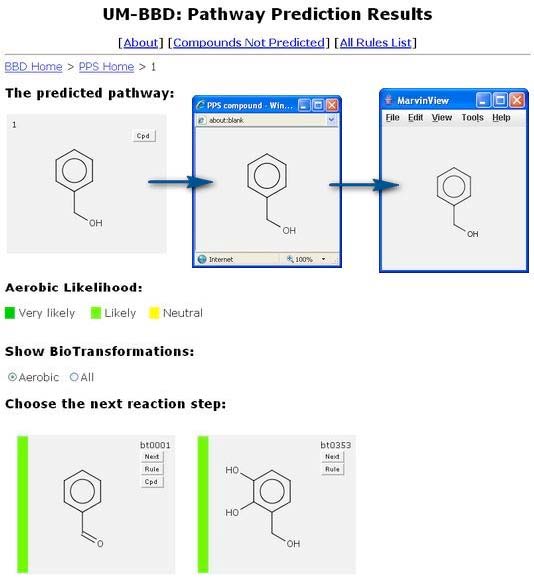
This will show the compound in a new window.
The compound can be rotated in this window. The window can also be increased in size, to better view larger compounds. Double clicking in this new window will produce a MarvinView window. The MarvinView window can provide much additional information on the compound. Information on how to use it is available at: http://www.chemaxon.com/marvin/chemaxon/marvin/help/view-index.html.
Aerobic Likelihood
Aerobic Likelihood is the likelihood that the reaction will occur under aerobic conditions, exposed to air, in soil (moderate moisture) or water, at neutral pH, 25°C, with no competing or toxic other compounds. Rules are assigned aerobic likelihood by two or more biodegradation experts. More information on the scoring process is available at:
http://umbbd.msi.umn.edu/predict/rulepriority.html
The aerobic likelihood of a compound is displayed as a vertical colored bar on the left side of a compound’s window. A color key is displayed beneath the “The Predicted Pathway.” The compounds are displayed in order of increasing aerobic likelihood.
A user is able to choose before or after a compound undergoes transformation if they will view all or only the aerobic likely transformations of a compound by selecting the appropriate button.
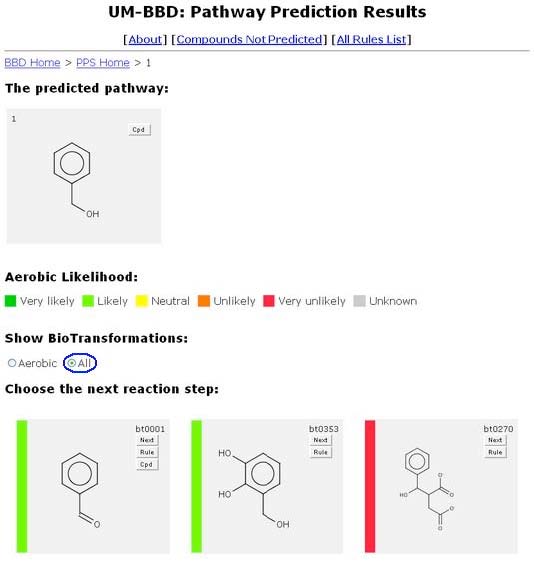
When the aerobic setting is used, an “immediate” feature is activated. All very likely and some likely rules are acted on immediately, without manual intervention. The prediction process appears to skip one or more steps. All steps are shown in the predicted pathway.
Rule Button
The PPS allows the user to click up to three buttons for each transformation. One of these, always shown, is the "Rule" button.
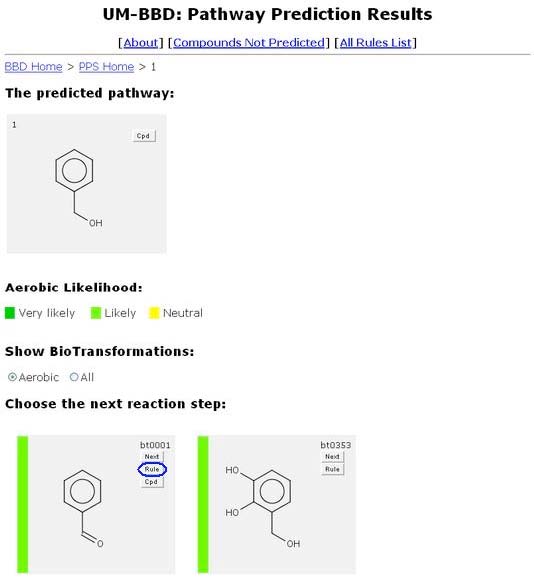
Clicking the "Rule" button will open a new browser window containing the rule page of the biotransformation rule associated with the specific reaction. A list of all rules the system uses is available. More information about rules is available in “About the BBD,” http://umbbd.msi.umn.edu/aboutBBD.html#btrule
Cpd Button
Another button is the "Cpd" button, located in the top right corner of an individual transformation beneath the "Rule" button. The "Cpd" button does not appear on every transformation, only on compounds found in the UM-BBD.
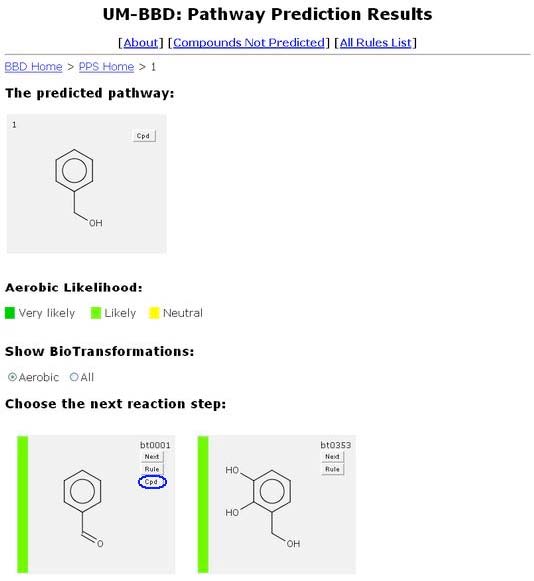
Clicking the "Cpd" button will open a new browser window containing the UM-BBD compound page for that compound. More information about compound pages is available in “About the BBD.”
http://umbbd.msi.umn.edu/aboutBBD.html#compound

Next Button
Another button that is always shown is the "Next" button. The "Next" button is located in the top right corner of each predicted compound’s graphic, above the "Rule" button.
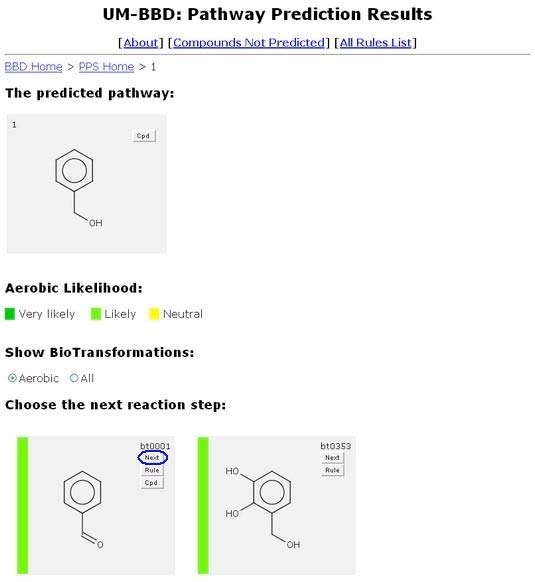
Clicking the "Next" button brings a user to the Pathway Prediction Results page for that transformation. The biotransformation the user selected will appear as the second step in the predicted pathway at the top of the page next to the original compound. The colored arrow indicates the aerobic likelihood of the biotransformation that connects the two compounds. Rule and Cpd buttons in predicted pathway graphics are the same as discussed under Results above.
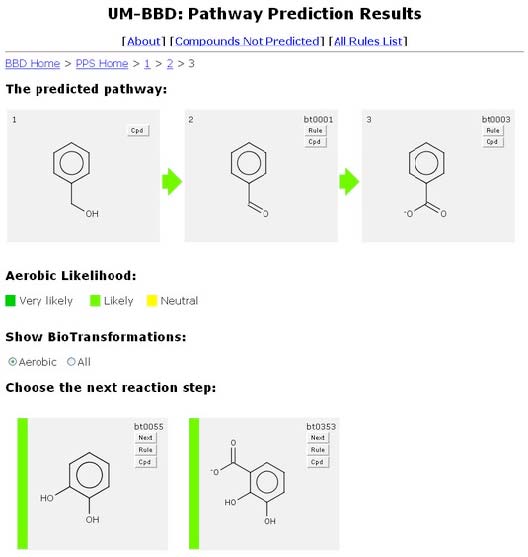
To continue the predicted pathway, select the "Next" button on another transformation. The Navigation Bar will the expand to “BBD Home > PPS Home > 1 > 2” for easy navigation to previous PPS prediction pages or BBD or PPS Home pages.
Green Next Button
After several cycles of the prediction process, you might see a green-shaded version of the “Next” button.
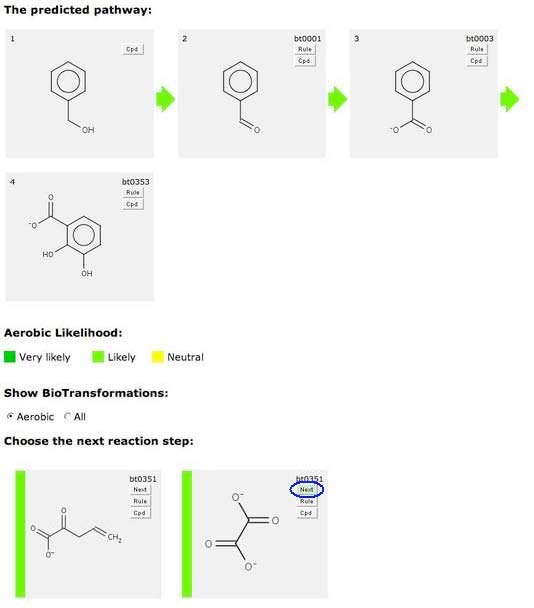
This indicates that this compound is readily degraded. If you click on a “Green Next” button, your predicted pathway is completed.
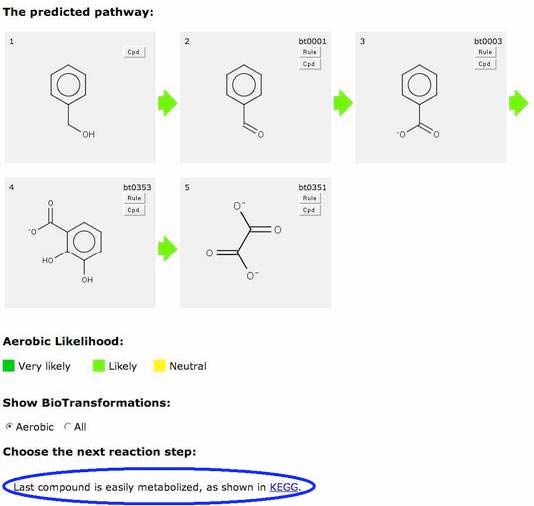
Alternatively, you may choose to continue prediction by clicking on a non-green “Next” button, if one is present.
Variable Aerobic Likelihood
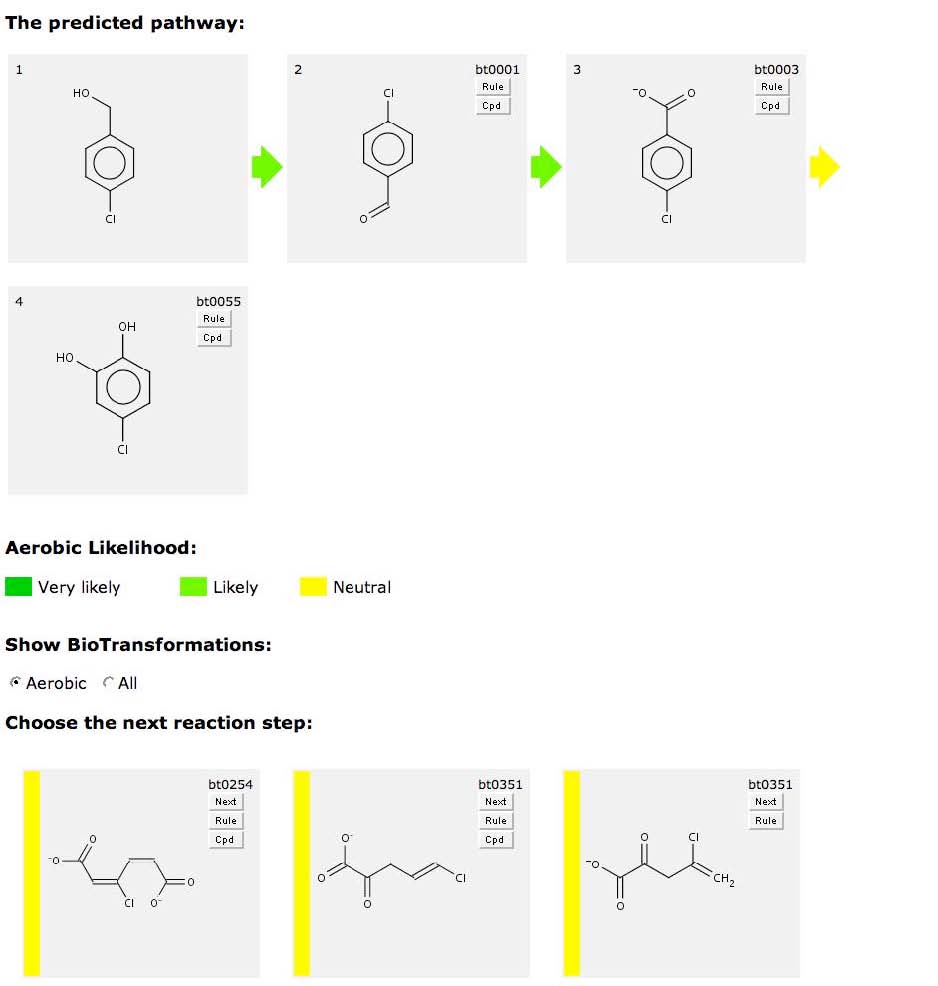
Some rules handle a wide range of compounds, and a single aerobiclikelihood is not appropriate for all of them. Thus structure-based variable aerobic likelihood has been added to the PPS. For aerobically likely aromatic ring monooxygenation, ringdioxygenation, ring cleavage, and ring decarboxylation rules, whenthe substrate contains one or more halogens, the aerobic likelihoodfor the transformation is changed to "neutral". This is the first steptoward better handling of aerobic likelihood.
Compare the previous predicted pathway for benzyl alcohol, withthe pathway below for p-chlorobenzyl alcohol. The aerobiclikelihoods for the first three transformations are likely, as above. However when the ring is oxygenated and cleaved, the aerobic likelihoods are neutral for the chlorinated compound, rather thanthe likely found for the non-halogen.
DRAWING STRUCTURES
The PPS uses Chemaxon's MarvinSketch for drawing compounds. Start from the PPS main page. To draw Benzyl Alcohol in the MarvinSketch window, select the "Benzene Ring" from the lower tool bar menu. Drag the structure to the desired location in the window and click to paste. Next select the "single bond" button from the left tool bar (for more options; e.g., double bond, or single bond up, click and hold the v to the right of the "single bond" button). With the "single bond" selected, click a corner of the benzene ring, drag the bond in the desired direction and release to paste. With the "single bond" still selected, click the methyl group, drag in the desired direction, and release to create a two carbon chain. To make the compound an alcohol, select the atomic symbol for oxygen, "O", from the right menu bar. Click on the methyl group to replace it with a hydroxide. A Marvin Sketch tutorial is available at http://www.chemaxon.com/jchem/marvin/chemaxon/marvin/help/about-sketch.html.
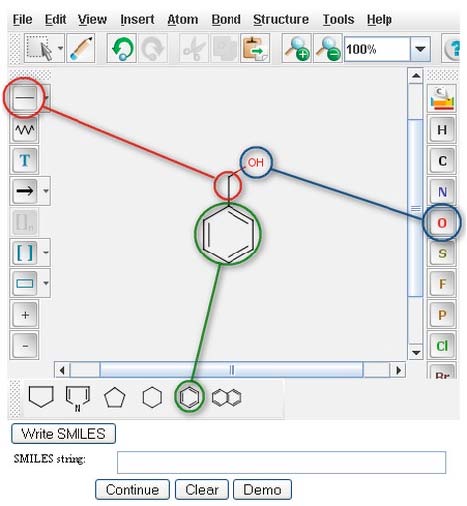
After creating an image image in the MarvinSketch window, a user next generates the SMILES string for that specific compound, by clicking the "Write SMILES" button located beneath the MarvinSketch window.
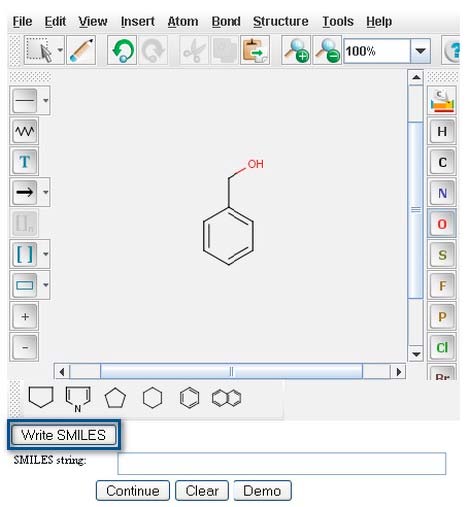
The SMILES string will appear in the “Smiles string:” window directly underneath your structure.
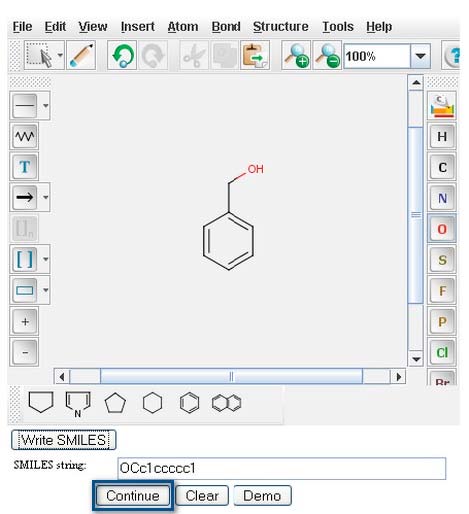
Click on the "Continue" button to proceed to “Pathway Prediction Results” for Benzyl Alcohol (page 4).
ENTERING SMILES STRINGS
If you know the SMILES string for your compound, you can instead enter a SMILES string directly. A SMILES string is an alphabetical representation of a compound. The SMILES string format is described on the SMILES Home Page from Daylight Chemical Information Systems, Inc.
http://www.daylight.com/smiles/
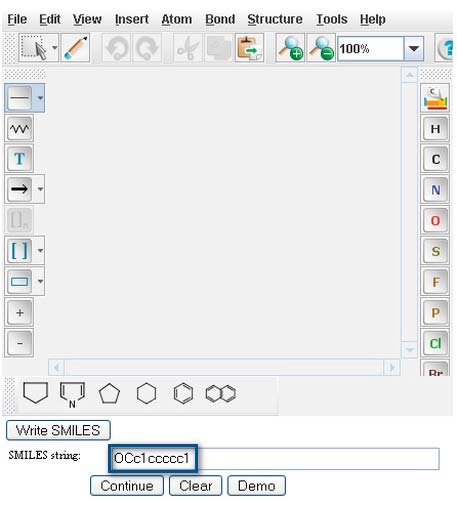
As a demonstration, type or paste the SMILES string of Benzyl Alcohol, OCc1ccccc1, into the "Smiles string" window.
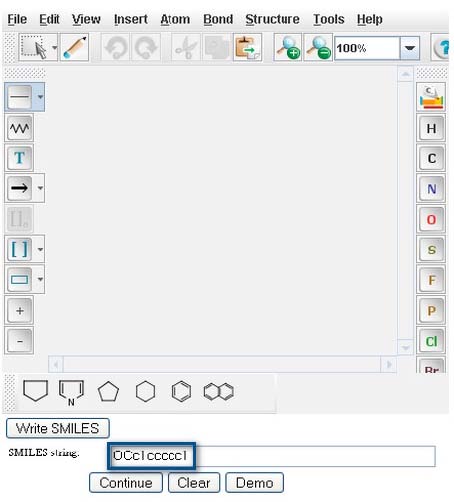
Click on the "Continue" button to proceed to “Pathway Prediction Results” for Benzyl Alcohol (page 4).
FOR FURTHER INFORMATION OR TO MAKE COMMENTS
To receive periodic progress reports, join the UM-BBD email list.
http://lists.umn.edu/cgi-bin/wa?SUBED1=um-bbd&A=1
Comments related to the database are appreciated. Our users are encouraged to contribute compound, reaction, and/or pathway information to the UM-BBD. To make comments or request more information “Contact Us”.
http://umbbd.msi.umn.edu/contact.html
BIBLIOGRAPHY
Ellis, L.B.M., Roe, D., Wackett, L.P. (2006) “The University of Minnesota Biocatalysis/Biodegradation Database: The First Decade” Nucleic Acids Research, 34: D517-D521.
Hou, B.K., Ellis, L.B.M., and Wackett, L.P. (2004) "Encoding Microbial Metabolic Logic: Predicting Biodegradation" Journal of Industrial Microbiology and Biotechnology, 31: 261-272.
Hou, B.K., Wackett, L.P., and Ellis, L.B.M. (2003) "Predicting Microbial Catabolism: A Functional Group Approach" Journal of Chemical Information and Computer Sciences, 43: 1051-1057.